20 years of sailing the biomolecular interaction landscape with HADDOCK

Guest post by Alexandre Bonvin (Utrecht University)
Reflecting on two decades of evolution, the HADDOCK (High Ambiguity Driven protein-protein DOCKing) integrative platform stands as a nuanced testament to the intricate landscape of biomolecular modelling. Developed at Utrecht University, this platform has quietly and steadily contributed to the understanding of biomolecular complexes. With its roots in pioneering research, HADDOCK has played and is still playing a crucial role in shaping the field of (computational) structural biology.
The 20 year anniversary symposium held in Huizen, Netherlands, on 7-10 November 2023, marked not just a celebration of achievements, but also an opportunity to delve into the platform’s ongoing significance since the publication of the first JACS paper in 2003.

HADDOCK in support of research and education
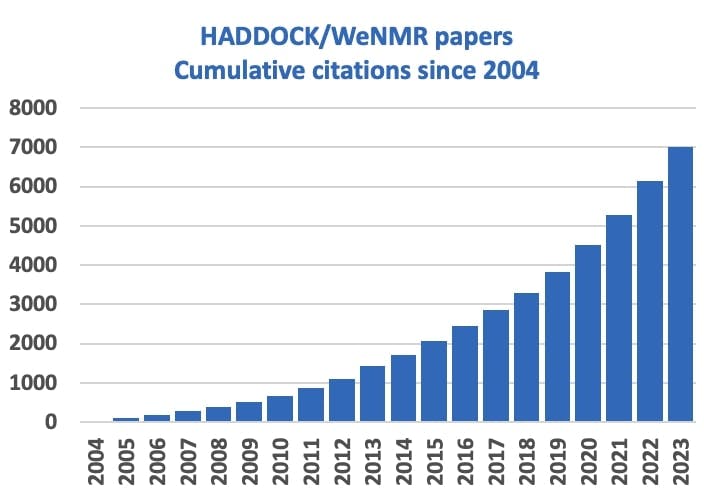
Since the original publication in 2003, HADDOCK has constantly evolved to provide support for more data sources and molecule types, from proteins, peptides, nucleic acids, to glycans and small ligands. Being easily accessible for non profit use through its web portal, HADDOCK has attracted a large and worldwide community of researchers. The original publications describing the various versions of HADDOCK and its web server have been cited more than 7000 times to date, reflecting its worldwide impact. Over the past 20 years we have also observed its embedding in educational programmes, seeing cohorts of students from various universities registering at given times throughout the year.
Currently, under BioExcel and in collaboration with the Netherlands eScience Center, HADDOCK is now evolving toward its third major version. This new modular and flexible implementation allows the creation of custom workflows, opening new opportunities for new scenarios and applications. This version, for which a front end web app is currently under development, will most likely evolve from using the EGI HTC resource to more cloud-based resources, again harvesting ideally the EGI/EOSC resources.
HADDOCK and EGI
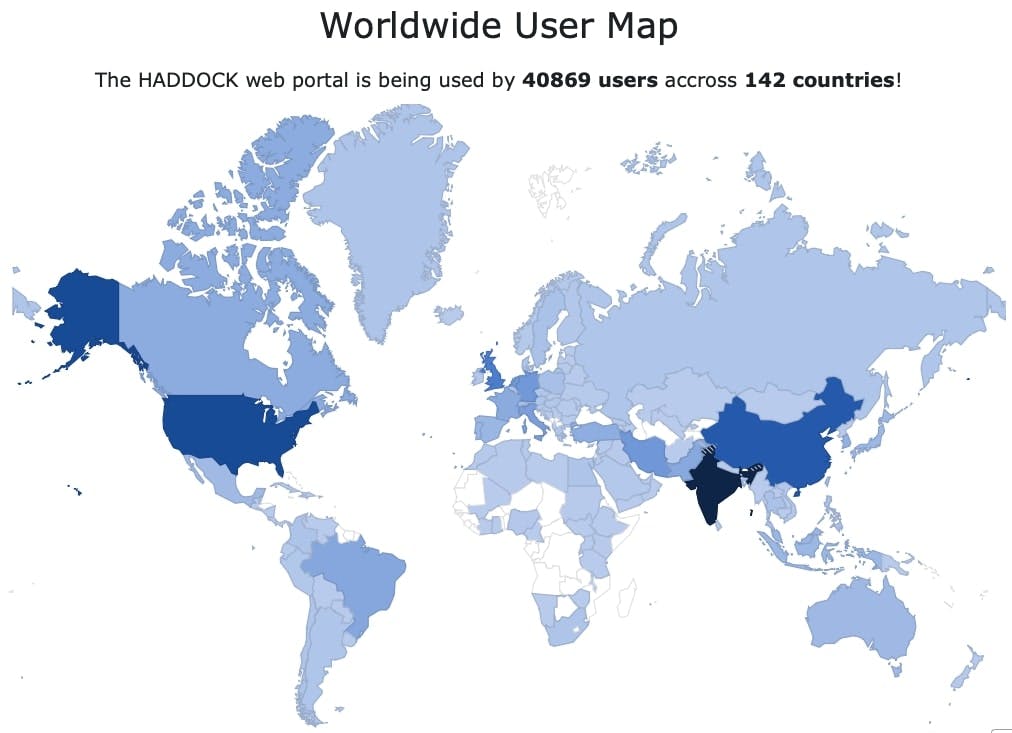
HADDOCK is an integrative platform for the modelling of biomolecular complexes which started making use of the EGI federation HTC resources since 2010 and has operated under a variety of EU projects over the years (eNMR, WeNMR, West-Life, EGI-Engage, EOSC-Hub, and EGI-ACE). The platform, offered as a web portal since 2008, is freely accessible to non-profit users, playing a crucial role in the WeNMR services and supported by the EGI Federation. With a thematic presence in the EOSC Marketplace, HADDOCK’s strategic approach has maximised its impact and valorization as a software tool. Boasting over 40,500 registered users from more than 142 countries, the portal provides not only application software but also automated pre- and post-processing, compute resources, temporary storage, and job scheduling and monitoring.
Notably, in the context of COVID-19 research, HADDOCK saw increased registrations, aligning with its role in the BioExcel Center of Excellence for Computational Biomolecular Research, committed to supporting COVID-19 researchers. The HADDOCK WeNMR team in Utrecht, collaborating with EGI and EOSC experts, enhanced processing capacity, including dedicated virtual clusters and customised solutions for researchers seeking tailored support.